Update cta_backtester.md
This commit is contained in:
parent
1d8e7c6897
commit
539e7aca1e
@ -102,8 +102,129 @@ def run_backtesting(
|
||||
|
||||
|
||||
## 参数优化
|
||||
vnpy提供2种参数优化的解决方案:穷举算法、遗传算法
|
||||
|
||||
参数优化功能使用的是穷举算法,即多进程对所有参数组合进行回测,并输出最终解集。其操作流程如下:
|
||||
|
||||
|
||||
|
||||
### 穷举算法
|
||||
|
||||
穷举算法原理:
|
||||
- 输入需要优化的参数名、优化区间、优化步进,以及优化目标。
|
||||
```
|
||||
def add_parameter(
|
||||
self, name: str, start: float, end: float = None, step: float = None
|
||||
):
|
||||
""""""
|
||||
if not end and not step:
|
||||
self.params[name] = [start]
|
||||
return
|
||||
|
||||
if start >= end:
|
||||
print("参数优化起始点必须小于终止点")
|
||||
return
|
||||
|
||||
if step <= 0:
|
||||
print("参数优化步进必须大于0")
|
||||
return
|
||||
|
||||
value = start
|
||||
value_list = []
|
||||
|
||||
while value <= end:
|
||||
value_list.append(value)
|
||||
value += step
|
||||
|
||||
self.params[name] = value_list
|
||||
|
||||
def set_target(self, target_name: str):
|
||||
""""""
|
||||
self.target_name = target_name
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
- 形成全局参数组合, 数据结构为[{key: value, key: value}, {key: value, key: value}]。
|
||||
```
|
||||
def generate_setting(self):
|
||||
""""""
|
||||
keys = self.params.keys()
|
||||
values = self.params.values()
|
||||
products = list(product(*values))
|
||||
|
||||
settings = []
|
||||
for p in products:
|
||||
setting = dict(zip(keys, p))
|
||||
settings.append(setting)
|
||||
|
||||
return settings
|
||||
```
|
||||
|
||||
|
||||
|
||||
- 遍历全局中的每一个参数组合:遍历的过程即运行一次策略回测,并且返回优化目标数值;然后根据目标数值排序,输出优化结果。
|
||||
```
|
||||
def run_optimization(self, optimization_setting: OptimizationSetting, output=True):
|
||||
""""""
|
||||
# Get optimization setting and target
|
||||
settings = optimization_setting.generate_setting()
|
||||
target_name = optimization_setting.target_name
|
||||
|
||||
if not settings:
|
||||
self.output("优化参数组合为空,请检查")
|
||||
return
|
||||
|
||||
if not target_name:
|
||||
self.output("优化目标未设置,请检查")
|
||||
return
|
||||
|
||||
# Use multiprocessing pool for running backtesting with different setting
|
||||
pool = multiprocessing.Pool(multiprocessing.cpu_count())
|
||||
|
||||
results = []
|
||||
for setting in settings:
|
||||
result = (pool.apply_async(optimize, (
|
||||
target_name,
|
||||
self.strategy_class,
|
||||
setting,
|
||||
self.vt_symbol,
|
||||
self.interval,
|
||||
self.start,
|
||||
self.rate,
|
||||
self.slippage,
|
||||
self.size,
|
||||
self.pricetick,
|
||||
self.capital,
|
||||
self.end,
|
||||
self.mode
|
||||
)))
|
||||
results.append(result)
|
||||
|
||||
pool.close()
|
||||
pool.join()
|
||||
|
||||
# Sort results and output
|
||||
result_values = [result.get() for result in results]
|
||||
result_values.sort(reverse=True, key=lambda result: result[1])
|
||||
|
||||
if output:
|
||||
for value in result_values:
|
||||
msg = f"参数:{value[0]}, 目标:{value[1]}"
|
||||
self.output(msg)
|
||||
|
||||
return result_values
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
注意:可以使用multiprocessing库来创建多进程实现并行优化。例如:若用户计算机是2核,优化时间为原来1/2;若计算机是10核,优化时间为原来1/10。
|
||||
|
||||
|
||||
|
||||
|
||||
穷举算法操作:
|
||||
|
||||
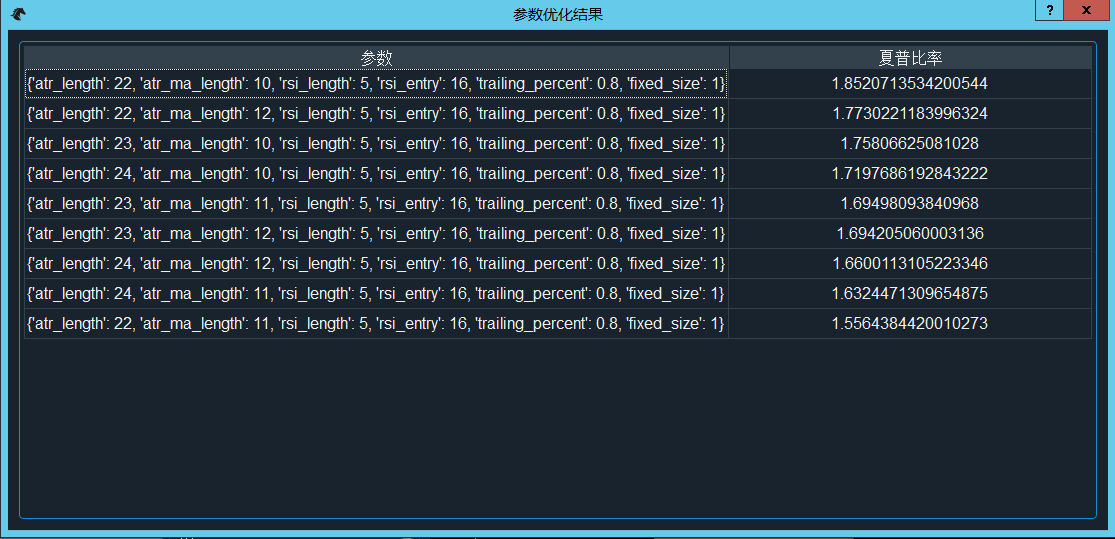
- 点击“参数优化”按钮,会弹出“优化参数配置”窗口,用于设置优化目标(如最大化夏普比率、最大化收益回撤比)和设置需要优化的参数以及优化区间,如图。
|
||||
|
||||
@ -118,6 +239,153 @@ def run_backtesting(
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
### 遗传算法
|
||||
|
||||
遗传算法原理:
|
||||
|
||||
- 输入需要优化的参数名、优化区间、优化步进,以及优化目标;
|
||||
|
||||
- 形成全局参数组合,该组合的数据结构是列表内镶嵌元组,即[[(key, value), (key, value)] , [(key, value), (key,value)]],与穷举算法的全局参数组合的数据结构不同。这样做的目的是有利于参数间进行交叉互换和变异。
|
||||
```
|
||||
def generate_setting_ga(self):
|
||||
""""""
|
||||
settings_ga = []
|
||||
settings = self.generate_setting()
|
||||
for d in settings:
|
||||
param = [tuple(i) for i in d.items()]
|
||||
settings_ga.append(param)
|
||||
return settings_ga
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
- 形成个体:调用random()函数随机从全局参数组合中获取参数。
|
||||
```
|
||||
def generate_parameter():
|
||||
""""""
|
||||
return random.choice(settings)
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
- 定义个体变异规则: 即发生变异时,旧的个体完全被新的个体替代。
|
||||
```
|
||||
def mutate_individual(individual, indpb):
|
||||
""""""
|
||||
size = len(individual)
|
||||
paramlist = generate_parameter()
|
||||
for i in range(size):
|
||||
if random.random() < indpb:
|
||||
individual[i] = paramlist[i]
|
||||
return individual,
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
- 定义评估函数:入参的是个体,即[(key, value), (key, value)]形式的参数组合,然后通过dict()转化成setting字典,然后运行回测,输出目标优化数值,如夏普比率、收益回撤比。(注意,修饰器@lru_cache作用是缓存计算结果,避免遇到相同的输入重复计算,大大降低运行遗传算法的时间)
|
||||
```
|
||||
@lru_cache(maxsize=1000000)
|
||||
def _ga_optimize(parameter_values: tuple):
|
||||
""""""
|
||||
setting = dict(parameter_values)
|
||||
|
||||
result = optimize(
|
||||
ga_target_name,
|
||||
ga_strategy_class,
|
||||
setting,
|
||||
ga_vt_symbol,
|
||||
ga_interval,
|
||||
ga_start,
|
||||
ga_rate,
|
||||
ga_slippage,
|
||||
ga_size,
|
||||
ga_pricetick,
|
||||
ga_capital,
|
||||
ga_end,
|
||||
ga_mode
|
||||
)
|
||||
return (result[1],)
|
||||
|
||||
|
||||
def ga_optimize(parameter_values: list):
|
||||
""""""
|
||||
return _ga_optimize(tuple(parameter_values))
|
||||
```
|
||||
|
||||
|
||||
|
||||
- 运行遗传算法:调用deap库的算法引擎来运行遗传算法,其具体流程如下。
|
||||
1)先定义优化方向,如夏普比率最大化;
|
||||
2)然后随机从全局参数组合获取个体,并形成族群;
|
||||
3)对族群内所有个体进行评估(即运行回测),并且剔除表现不好个体;
|
||||
4)剩下的个体会进行交叉或者变异,通过评估和筛选后形成新的族群;(到此为止是完整的一次种群迭代过程);
|
||||
5)多次迭代后,种群内差异性减少,整体适应性提高,最终输出建议结果。该结果为帕累托解集,可以是1个或者多个参数组合。
|
||||
|
||||
注意:由于用到了@lru_cache, 迭代中后期的速度回提高非常多,因为很多重复的输入都避免了再次的回测,直接在内存中查询并且返回计算结果。
|
||||
```
|
||||
from deap import creator, base, tools, algorithms
|
||||
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
|
||||
creator.create("Individual", list, fitness=creator.FitnessMax)
|
||||
......
|
||||
# Set up genetic algorithem
|
||||
toolbox = base.Toolbox()
|
||||
toolbox.register("individual", tools.initIterate, creator.Individual, generate_parameter)
|
||||
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
|
||||
toolbox.register("mate", tools.cxTwoPoint)
|
||||
toolbox.register("mutate", mutate_individual, indpb=1)
|
||||
toolbox.register("evaluate", ga_optimize)
|
||||
toolbox.register("select", tools.selNSGA2)
|
||||
|
||||
total_size = len(settings)
|
||||
pop_size = population_size # number of individuals in each generation
|
||||
lambda_ = pop_size # number of children to produce at each generation
|
||||
mu = int(pop_size * 0.8) # number of individuals to select for the next generation
|
||||
|
||||
cxpb = 0.95 # probability that an offspring is produced by crossover
|
||||
mutpb = 1 - cxpb # probability that an offspring is produced by mutation
|
||||
ngen = ngen_size # number of generation
|
||||
|
||||
pop = toolbox.population(pop_size)
|
||||
hof = tools.ParetoFront() # end result of pareto front
|
||||
|
||||
stats = tools.Statistics(lambda ind: ind.fitness.values)
|
||||
np.set_printoptions(suppress=True)
|
||||
stats.register("mean", np.mean, axis=0)
|
||||
stats.register("std", np.std, axis=0)
|
||||
stats.register("min", np.min, axis=0)
|
||||
stats.register("max", np.max, axis=0)
|
||||
|
||||
algorithms.eaMuPlusLambda(
|
||||
pop,
|
||||
toolbox,
|
||||
mu,
|
||||
lambda_,
|
||||
cxpb,
|
||||
mutpb,
|
||||
ngen,
|
||||
stats,
|
||||
halloffame=hof
|
||||
)
|
||||
|
||||
# Return result list
|
||||
results = []
|
||||
|
||||
for parameter_values in hof:
|
||||
setting = dict(parameter_values)
|
||||
target_value = ga_optimize(parameter_values)[0]
|
||||
results.append((setting, target_value, {}))
|
||||
|
||||
return results
|
||||
```
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
Loading…
Reference in New Issue
Block a user